1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69 | ############################
# Generate best estimate for frequency and amplitude given irregular timeseries
# displaying oscillations by fitting a sinusoidal model.
############################
### Define sinusoid model
mod=function(A,f,c,p,t) A*sin(2*pi*f*t+p)+c
### Use model to generate a synthetic irregular time-series (with noise)
### For a real problem, replace this section with actual data!
# True paramter values
Atrue=5; ftrue=1/30; ctrue=20; ptrue=0
# Random simulation of irregular observation schedule
expt=sort(sample(0:100,50,replace=FALSE))
# Generate realistic dataset (irregular observation times and observations with error)
df=data.frame(time=expt,obs=mod(Atrue,ftrue,ctrue,ptrue,expt)+rnorm(length(expt),mean=1,sd=2))
### Estimate frequency of data by simulating a regular time-series
# Construct interpolating function for observed data
intf=approxfun(df$time,df$obs)
# Simulate a regular measurement frequency
mfreq=2
# Create regularly spaced time-series
regtimes=min(df$time)+mfreq*(0:((max(df$time)-min(df$time))/mfreq))
regobs=intf(regtimes)
# Generate and plot frequency spectrum for regular time-series
spec=spectrum(regobs,plot=TRUE)
# Extract the fundamental frequency
fguess=spec$freq[which.max(spec$spec)]
# Convert units from measurent frequency original units
fguess=fguess/mfreq
### Estimate other paramters
# Estimate amplitude as half range of observations
Aguess=(max(df$obs)-min(df$obs))/2
# Estimate offset as mean of observations
cguess=mean(df$obs)
# Estimate the phase as
pguess=asin((df$obs[1]-cguess)/Aguess)-2*pi*fguess*df$time[1]
### Fit model to data
# Objective function (sum of squared difference between model and observations)
obj<-function(z){
A=z[1]; f=z[2]; c=z[3]; p=z[4];
return(sum((mod(A,f,c,p,df$time)-df$obs)**2))
}
# Optimisation
# Approximate sampling frequency (to avoid Nyquist limit)
sfreqest=length(df$time)/max(df$time)
lowerlim=c(0,0,-Inf,0)
upperlim=c(Inf,2*sfreqest,Inf,2*pi)
params=optim(c(Aguess,fguess,cguess,pguess),obj,lower=lowerlim,upper=upperlim,method="L-BFGS-B",control=list(maxit=10000))$par
Aopt=params[1]; fopt=params[2]; copt=params[3]; popt=params[4]
Apercent=100*(Atrue-Aopt)/Atrue; fpercent=100*(ftrue-fopt)/ftrue; cpercent=100*(ctrue-copt)/ctrue; ppercent=100*(ptrue-popt)/(2*pi)
report=paste("% Diffs: A:",signif(Apercent,3),"f:",signif(fpercent,3),"c:",signif(cpercent,3),"p:",signif(ppercent,3))
# Inspect goodness of fit
#pdf("Oscillations.pdf",width=8,height=8)
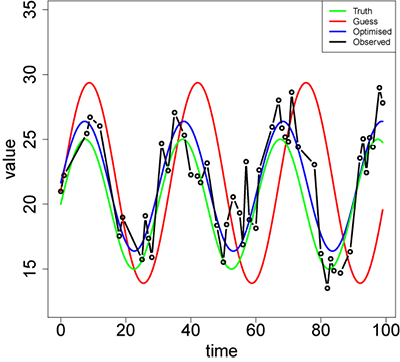
plot(NULL,,xlim=c(min(df$time),max(df$time)),ylim=c(0.9*min(df$obs),1.2*max(df$obs)),type="n",xlab="time",ylab="value",main=report,cex.lab=2,cex.axis=2)
curve(mod(Aguess,fguess,cguess,pguess,x),min(df$time),max(df$time),add=TRUE,col="red",lwd=3,,n=1001)
curve(mod(Aopt,fopt,copt,popt,x),min(df$time),max(df$time),add=TRUE,col="blue",lwd=3,n=1001)
curve(mod(Atrue,ftrue,ctrue,ptrue,x),min(df$time),max(df$time),add=TRUE,col="green",lwd=3,n=1001)
points(df$time,df$obs,type="b",lwd=3)
legend("topright",c("Truth","Guess","Optimised","Observed"),lwd=3,col=c("green","red","blue","black"))
#dev.off()
# Generate a small report
res=data.frame(A=c(Atrue,Aguess,Aopt),f=c(ftrue,fguess,fopt),c=c(ctrue,cguess,copt),p=c(ptrue,pguess,popt))
rownames(res)=c("Truth","Guess","Optimised")
print(res)
|