Conor Lawless
Scientific Computing
data science, dynamic simulation modelling, genomics, interactive visualisation, dashboards, image & video analysis
e: cnr.lwlss@gmail.comt: @cnrlwlss
About me
Sampling from skeletal muscle sections by laser capture microdissection
What proportion of fibres can we dissect from a section before it starts to disintegrate?
Mitochondria are the parts of our cells that are responsible for generating energy. In the Wellcome Centre for Mitochondrial Research, we try to understand ways in which the function of mitochondria deteriorates in individual cells during the progression of mitochondrial diseases. There is a good introduction to mitochondrial biology and mitochondrial diseases in this blog post by Max Piotrowicz.
An important component of the work in the centre is analysing skeletal muscle sections, which are thin, cross-sectional slices across hundreds of long, post-mitotic muscle fibres, which are packed very closely together in muscle tissue. These sections are usually sampled from patient muscle biopsies. Muscle fibres are the cells that allow us to move and are among the most energy-demanding cell type in our bodies. As a result of their high energy requirement, these cells are particularly vulnerable to the mitochondrial dysfunction, known as myopathy, that arises during mitochondrial disease.
In a single section, it is possible to observe a mosaic pattern of respiratory chain deficiency: there is often little or no correlation between how dysfunctional mitochondria are in one fibre and how dysfunctional they are in its neighbours. This random, mosaic pattern gives us a clue that the progression of mitochondrial disease is stochastic and that stochastic progression occurs independently in all cells.
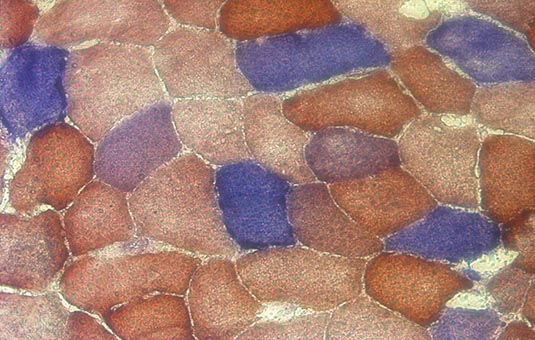 Microscopy image showing mosaic patterin in part of a histological cross-section of skeletal muscle. Cells stained blue are COX-negative, indicating mitochondrial dysfunction.
Microscopy image showing mosaic patterin in part of a histological cross-section of skeletal muscle. Cells stained blue are COX-negative, indicating mitochondrial dysfunction.
There are many types of measurements that we can make on individual fibres within a muscle section, but typically, before analysis of sections at the scale of single cells, we need to be able to identify the location of individual fibres, either computationally or using the experience and judgement of an expert. A common approach is to capture an image of the section (e.g. a fluorescence microscopic image, highlighting the expression of proteins of interest in each cell), to then use image analysis techniques to identify the location of fibres and then to summarise protein expression from colour intensities at each location in the image.
Followup experiments can be carried out where we physically cut the section apart and extract material from individual fibres. Laser capture microdissection, where the edges of cells are identified and cut out using a high-powered laser connected to a microscope, is one way to very precisely dissect material from individual fibres and we often use this method to access material from single cells for detailed DNA or RNA sequencing analysis.
One major difficulty with laser microdissection is that, when you cut a fibre from a section, it weakens the section considerably at that location. If you dissect away too many adjacent fibres, at some point, the section will collapse or disintegrate. This is not a problem if you just want to sample a few single fibres from a section, you can simply choose fibres that are far apart. However, the partially destructive nature of this approach limits the proportion of fibres that can be cut from a section.
This post is to estimate what proportion of fibres we can reliably microdissect from a skeletal muscle section before the section begins to fall apart. This should give us an idea about the maximum proportion of fibres that we can reliably sample from a section in order to take material forward for further analysis. I will assume that we have already identified the location (the x- and y-coordinates of the centres) and size (area) of each fibre. The image analysis software mitocyto can help with locating and measuring fibres.
If we assume that we have to leave the immediate neighbours of every sampled fibre intact, how many fibres does that allow us to sample? The first problem is to identify the neighbours of each fibre. To do that, I use Delaunay triangulation to calculate an adjacency matrix for all of the fibre locations in the image. In order to account correctly for possible holes in the section, I do not define any of the Delaunay neighbours whose distance apart is more than five times the radius estimated from the list of fibre areas in the input (assuming circular fibres) as adjacent. I then randomly select a fibre for sampling from all available fibres, classify all adjacent fibres as unavailable for subsequent sampling and then repeat until all fibres are either sampled or unavailable. Some R code for carrying out calculations and generating plots here.
 Animation showing sampling strategy when immediate neighbours of sampled fibres become unavailable for subsequent micro-dissection. Left panel shows Delaunay triangulation from list of fibre locations. Right panel shows raw image data. Yellow dots overlaid indicate measured fibre centres. Red dots are fibres selected for micro-dissection. Blue dots are fibres which are unavailable due to being neighbours of sampled fibre.
Animation showing sampling strategy when immediate neighbours of sampled fibres become unavailable for subsequent micro-dissection. Left panel shows Delaunay triangulation from list of fibre locations. Right panel shows raw image data. Yellow dots overlaid indicate measured fibre centres. Red dots are fibres selected for micro-dissection. Blue dots are fibres which are unavailable due to being neighbours of sampled fibre.
We can see that, after going through the procedure outlined above once for the example image, the highest proportion that we can sample while maintaining the structural integrity of the tissue is 26.2%. This example suggests that we cannot sample high proportions of fibres by laser microdissection.
If we wanted to be more conservative we could insist that, as well as the neighbours of a sampled fibre becoming unavailable, that their neighbours also became unavailable. In this way we would ensure that there are at least two fibres between sampled locations, which should result in a more stable tissue throughout sampling.
 Animation showing sampling strategy when both immediate neighbours of sampled fibres and their neighbours become unavailable for subsequent micro-dissection. Colours as above.
Animation showing sampling strategy when both immediate neighbours of sampled fibres and their neighbours become unavailable for subsequent micro-dissection. Colours as above.
Taking this second neighbour approach we can see that the maximum proportion that can be sampled in this example has reduced to 11.1%.
In practice I note that the maximum proportion of fibres that I’ve seen sampled from muscle sections is ~10%.
This analysis suggests to me that, using laser capture micro-dissection, the proportion of fibres that can be reliably sampled from a skeletal muscle section is between 10-25% of the total. This low proportion represents a fairly heavy constraint on the design of single-cell experiments based on laser micro-dissection. In particular, complete sampling is not possible. The main advantage of complete sampling is that it is completely unbiased. However we can still achieve unbiased, partial sampling by using a computer to randomly select fibres for dissection.