In part I of this series of posts, I presented the maths behind posing circle packing as a high-dimensional, non-linear, constrained optimisation problem. I am using this problem as an example to try out various optimisation algorithms. In part II I demonstrated an implementation of this problem in R, together with an R function for visualising candidate solutions.
In this post, I will describe some findings from attempts to optimise the circle packing objective function with R's default, general-purpose optimisation package: optim and two other packages for performing global optimisation using genetic algorithms with derivative-based intermediate searches.
The code below requires the R functions from part II, included in the CirclePacking.R script which you can download here.
I've written a second script demonstrating the use of the functions in CirclePacking.R in combination with the 3 optimisation packages mentioned above (see the bottom of this post). Running this code generates plots like the one below, visualising solutions, together with some statistics about running time distributions and packing density distributions for ensembles of solutions.
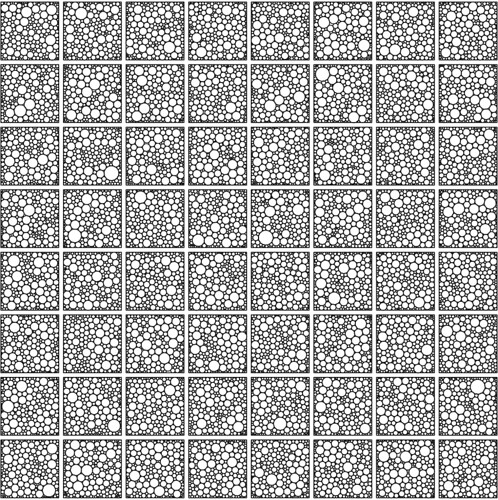
64 solutions for packing 200 circles into a square with the optim package.
The optim package includes an implementation of the L-BFGS-B algorithm, which is an efficient, derivative-based local optimiser allowing lower and upper bounds to be placed on parameters.
The rgenoud package uses genetic algorithms and derivatives to solve global optimisation problems. I used it successfully to fit a mixture model to pixel intensity distributions as part of the segmentation step in the image analysis tool: Colonyzer. It also has some interesting functionality for distributing computing load across multiple CPUs, by using a snow cluster which I wanted to investigate.
The DEoptim package also uses genetic algorithms and derivatives to solve global optimisation problems. I have previously found that this algorithm was faster than rgenoud when automatically fitting a logistic model to yeast growth curves in Quantitative Fitness Analysis.
Results
My findings are problem-specific, and depend on the number of circles allowed (which you can vary by modifying the code below) but so far, they are as follows:
- For all N I've tried (up to 200), the L-BFGS-B algorithm was fastest. It achieved the same packing densities, but in shorter times.
- Using the rgenoud cluster functionality did not speed up the genoud algorithm, or improve the packing densities of its solutions.
- DEoptim crashes with mysterious errors for N>200 (more than 600 parameters) on a 64-bit machine with 12 Gb RAM. However, this is probably many more parameters than are required for most practical purposes.
- rgenoud was much faster than DEoptim in this case (achieved the same or higher packing densities more quickly).
It's difficult to draw solid conclusions from this analysis. Result 1) was particularly surprising since I expected this high-dimensional problem to be very vulnerable to getting stuck in local minima, and so I expected global optimisers to be more efficient. As mentioned above, result 4) is reversed for at least one other optimisation problem.
However, I'm happy with the solutions (see image above), they have very pleasing organic patterns.